Imprinting.ppt
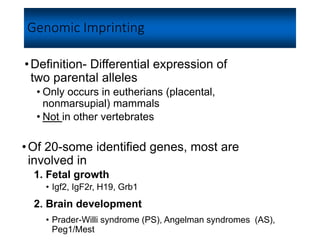
- 1. Genomic Imprinting •Definition- Differential expression of two parental alleles • Only occurs in eutherians (placental, nonmarsupial) mammals • Not in other vertebrates •Of 20-some identified genes, most are involved in 1. Fetal growth • Igf2, IgF2r, H19, Grb1 2. Brain development • Prader-Willi syndrome (PS), Angelman syndromes (AS), Peg1/Mest
- 6. Genomic-imprinting conflict What is genomic imprinting and why has it evolved? Expression of a gene depending on whether inherited from father or mother Main arenas of imprinting effects on human health: -Placenta -Brain -Carcinogenesis -Stem cells -In vitro fertilization from dad from mom
- 7. • In experiments on mice maternal and paternal pronuclei normal mouse both paternal pronuclei The placenta is huge, the embryo is undeveloped both matern al pronucl ei The embryo is formed, the placenta is underdeveloped
- 8. A summary of imprinted gene functions during embryogenesis. Robert N. Plasschaert, and Marisa S. Bartolomei Development 2014;141:1805-1813 © 2014. Published by The Company of Biologists Ltd
- 9. CONFLICT THEORY OF IMPRINTING -> abundant support from empirical studies of imprinted genes and growth, in mice and humans (1) IGF2-IGF2R (Haig & Graham 1991 Cell) (2) CDKN1C (Andrews et al. 2007 BMC Dev Biol) (3) GRB10 (Charalambous et al. 2003 PNAS) Beckwith- Wiedemann Syndrome Silver- Russell syndrome Mighty mouse Normal sized human 2 doses IGF2 1 dose IGF2 0 doses IGF2
- 10. Genomic imprinting the unequal expression of the maternal and paternal alleles of a gene XX X Y Maternal allele Patternal allele Patternal allele Matternal allele
- 11. • Two major clusters of imprinted genes have been identified in humans, one on the short (p) arm of chromosome 11 (at position 11p15) and another on the long (q) arm of chromosome 15 (in the region 15q11 to 15q13).
- 12. Mouse models to study genomic imprinting that allow the maternal and paternal chromosome to be distinguished. Denise P. Barlow, and Marisa S. Bartolomei Cold Spring Harb Perspect Biol 2014;6:a018382 ©2014 by Cold Spring Harbor Laboratory Press Mouse models to study genomic imprinting that allow the maternal and paternal chromosome to be distinguished. Mammals are diploid and inherit a complete chromosome set from the maternal and paternal parent. However, mice can be generated that (1) inherit two copies of a chromosome pair from one parent and no copy from the other parent (known as UPD), (2) inherit a partial chromosomal deletion from one parent and a wild-type chromosome from the other parent, and (3) inherit chromosomes carrying single- nucleotide polymorphisms (known as SNPs) from one parent and a wild-type chromosome from the other parent. Offspring with UPDs or deletions are likely to display lethal phenotypes, whereas SNPs will allow the production of viable offspring.
- 13. A maternal and paternal genome are needed for mammalian reproduction. Denise P. Barlow, and Marisa S. Bartolomei Cold Spring Harb Perspect Biol 2014;6:a018382 ©2014 by Cold Spring Harbor Laboratory Press A maternal and paternal genome are needed for mammalian reproduction. The nuclear transfer technique used micropipettes and high-powered microscopes to remove the male or female nuclei from a newly fertilized egg and place them in various combinations into a second “host” fertilized egg that had already been enucleated, thereby generating anew diploid embryos with two maternal (gynogenetic) or two paternal (androgenetic) genomes or a biparental genome (wild-type). Gynogenetic and androgenetic embryos were lethal at early embryonic stages. Only reconstituted embryos that received both a maternal and paternal nucleus (wild-type) survived to produce living young. These experiments show the necessity for both the maternal and paternal genome in mammalian reproduction, and indicate the two parental genomes express different sets of genes needed for complete embryonic development.
- 14. Imprint acquisition and erasure in mammalian development. Denise P. Barlow, and Marisa S. Bartolomei Cold Spring Harb Perspect Biol 2014;6:a018382 ©2014 by Cold Spring Harbor Laboratory Press Imprint acquisition and erasure in mammalian development. Imprints are acquired by the gametes; thus, oocytes and sperm already carry imprinted chromosomes (first-generation imprints). After fertilization when the embryo is diploid, the imprint is maintained on the same parental chromosome after each cell division in cells of the embryo, yolk sac, placenta, and also in the adult. The germ cells are formed in the embryonic gonad and the imprints are erased only in these cells before sex determination. As the embryo develops into a male, the gonads differentiate to testes that produce haploid sperm that acquire a paternal imprint on their chromosomes. Similarly, in developing females, chromosomes in the ovaries acquire maternal imprints (second-generation imprints).
- 15. Imprinted genes play a role in mammalian reproduction. Denise P. Barlow, and Marisa S. Bartolomei Cold Spring Harb Perspect Biol 2014;6:a018382 ©2014 by Cold Spring Harbor Laboratory Press Imprinted genes play a role in mammalian reproduction. Mammals are diploid and reproduction requires fertilization of a haploid female egg by a haploid male sperm to recreate a diploid embryo. Only females are anatomically equipped for reproduction, but they cannot use parthenogenesis to reproduce (the possibility of which is represented by a pink dashed line) because essential imprinted genes needed for fetal growth are imprinted and silenced on maternal chromosomes. These genes are expressed only from paternal chromosomes; thus, both parental genomes are needed for reproduction in mammals. Parthenogenesis is the production of diploid offspring from two copies of the same maternal genome.
- 16. Imprinted genes are expressed from one parental allele and often clustered. Denise P. Barlow, and Marisa S. Bartolomei Cold Spring Harb Perspect Biol 2014;6:a018382 ©2014 by Cold Spring Harbor Laboratory Press Imprinted genes are expressed from one parental allele and often clustered. Most imprinted genes (yellow) are found in clusters that include multiple protein-coding mRNAs (IG) and at least one noncoding RNA (IG-NC). Nonimprinted genes can also be present (NI in gray). The imprinting mechanism is cis acting and imprinted expression is controlled by an imprint control element (ICE) that carries an epigenetic imprint inherited from one parental gamete.
- 17. Imprinted expression is regulated by gametic DMRs (G-DMR). Denise P. Barlow, and Marisa S. Bartolomei Cold Spring Harb Perspect Biol 2014;6:a018382 ©2014 by Cold Spring Harbor Laboratory Press Imprinted expression is regulated by gametic DMRs (G-DMR). (Left) The effect of deleting the gametic DMR from the imprinted chromosome (green). (Right) The effect of deleting the G-DMR from the nonimprinted chromosome (yellow). In many imprinted clusters (e.g., Igf2r, Kcnq1, and Dlk1), experimental deletion of the G-DMR only affects the chromosome carrying the nonimprinted G-DMR. This results in a loss of repression of the imprinted protein-coding mRNA genes (IG) and a gain of repression of the imprinted lncRNA gene (IG-NC). Note that in some imprinted clusters (Igf2 and Pws) that are not illustrated here, the methylated G-DMR appears also to be required for expression of some of the imprinted mRNAs in cis. del, deleted DNA; G-DMR, gametic differentially DNA-methylated region; NG, nonimprinted gene; arrow, expressed allele; slashed circle, repressed allele; imprint, epigenetic modification leading to a change in gene expression in cis.
- 18. Two cis-acting silencing mechanisms at imprinted gene clusters. Denise P. Barlow, and Marisa S. Bartolomei Cold Spring Harb Perspect Biol 2014;6:a018382 ©2014 by Cold Spring Harbor Laboratory Press Two cis-acting silencing mechanisms at imprinted gene clusters. (A) Insulator model for the Igf2 cluster. The expression pattern for endoderm is shown. On the maternal chromosome, the unmethylated ICE binds the CTCF protein and forms an insulator that prevents the common endoderm enhancers (E) from activating Igf2 and Ins2. Instead the enhancers activate the nearby H19 lncRNA promoter. On the paternal chromosome, the methylated ICE cannot bind CTCF and an insulator does not form; hence the Igf2 and Ins2 mRNA genes are expressed only on this chromosome. The H19 lncRNA is methylated, most likely because of spreading from the 2-kb distant methylated ICE, and silenced.
- 19. Two cis-acting silencing mechanisms at imprinted gene clusters. Denise P. Barlow, and Marisa S. Bartolomei Cold Spring Harb Perspect Biol 2014;6:a018382 ©2014 by Cold Spring Harbor Laboratory Press Two cis-acting silencing mechanisms at imprinted gene clusters. (B) lncRNA model for the Igf2r cluster. The expression pattern for placenta is shown. On the maternal chromosome, the methylated ICE contains the Airn lncRNA promoter that is directly silenced by the DNA methylation imprint. The Igf2r, Slc22a2, and Slc22a3 mRNA genes are expressed only on this chromosome. Mas1 and Slc22a1 are not expressed in placenta (filled diamond). On the paternal chromosome, the Airn lncRNA promoter lying in the unmethylated ICE is expressed and silences Igf2r (in part by kicking off RNA polymerase II), Slc22a2, and Slc22a3 in cis. Note that in both models, the DNA methylation imprint silences the lncRNA and permits mRNA expression. ICE, imprint control element; gray arrow, expressed allele of an imprinted gene; slashed circle, repressed allele of an imprinted gene; thick gray arrows, long distance effect in cis.
- 20. Paternally expressed imprinted genes that function as growth promoters (i.e., Igf2, Peg1, Peg3, Rasgrf1, Dlk1) and show growth retardation in embryos deficient for the gene. There are also maternally expressed imprinted genes that function as growth repressors (i.e., Igf2r, Gnas, Cdkn1c, H19, Grb10), as shown by a growth enhancement in embryos deficient for the gene.
- 21. Categories of imprinted genes 1. Fetal growth genes- Insulin-like growth factor-like II (IGF2) response pathway • IGF2 • Igf2r • Grb10 • H19 • Gnas • Rasgrf • Mash2 • Why?- Embryo develops in a parasite-like relationship with mother.
- 22. Categories of imprinted genes 2. Brain development- • Prader Willi Syndrome- paternal chromosome deletion • Angelman Syndrome- maternal chromosome deletion
- 23. Prader-Willi Syndrome • 1 in 15,000 live births • mostly sporatic • deletion at 15 q11-q13 • diagnosis at 2 years • compulsive overeaters
- 24. A typical Prader-Willi patient Prader Willi Syndrome- Due to paternal chromosome deletion
- 25. • 1 in 25,000 live births • mostly sporatic • 80 % have deletion at 15q11-q13 • Specifically mutation of UBE3A gene Angelman Syndrome
- 26. Angelman Syndrome •Speech impairment •None or minimal use of words •Receptive and non-verbal communication skills higher than verbal ones •Movement or balance disorder, usually ataxia of gait •Behavioral uniqueness: any combination of frequent laughter/smiling; apparent happy demeanor •easily excitable personality, often with hand flapping movements; hypermotoric behavior; short attention span
- 27. Angelman Syndrome Normal –Angelman Syndrome- maternal chromosome deletion Or 2 paternal chromosomes Or an imprinting defect
- 28. Parent Offspring Conflict Hypothesis (Haig hypothesis) • Conflict between male and female over allocation of maternal resources to offspring • Dad uses imprinting to direct all resources to immediate offspring (not future litters) • Mom uses imprinting to allocate resources to multiple litters • Thus, predict paternally expressed genes would promote growth, maternally expressed genes should slow it down • Prediction mostly hold true • Example- Igf2 (paternally expressed)-if defective=40% reduction in growth
- 29. Parent Offspring Conflict Hypothesis (Haig hypothesis) Example – The Igf2 gene and its receptor Igf2r •Igf2 (paternally expressed)-if defective=40% reduction in growth •Igf2r (Igf2 receptor)- if defective=increase growth •Igf2-/Igf2r- = normal Another test- Ask if imprinting fails to occur in a monogomous species The Beach mouse is entirely monogomous ….but imprinting still occurs, contrary to model
- 30. How are imprinted genes silenced? S. Tilghman, Cell 96:185
- 31. How are imprinted genes silenced? S. Tilghman, Cell 96:185 Dnmt-/- mice- Many imprinted genes (e.g. H19) reactivated ..but, Igf2 and Igf2r are silenced Mechanism- Methylation interferes with transcription factor binding Problems with model- 1. Promoters of silent Igf2 and Igf2r alleles are unmethylated 2. One gene, Mash2, is unaffected by loss of methylation
- 32. How are imprinted genes silenced? S. Tilghman, Cell 96:185 Mechanism- Promoters compete for a single enhancer Problem with model- Both H19 and Igf2 are expressed if H19 gene replaced with luciferase Igf2 H19
- 33. How are imprinted genes silenced? S. Tilghman, Cell 96:185 Epigenetic marker binds to unmethylated DNA Mechanism- Methylation serves two purposes 1. Inactivate a gene (e.g. H19) 2. Prevent binding of epigenetic marker so that Igf2 is activated Igf2 H19 Epigenetic insulator prevents enhancer from “talking to” Igf2 Evidence in support: if delete insulator element- both Igf2 and H19 expressed
- 34. Evidence for chromatin boundary mechanism Deletion of ICR- both genes expressed Identify protein (called CTCF) that binds ICR CTCF cannot bind methylated DNA Thorvaldsen and Bartolmei, Science 288:2145, 2000
- 35. How are imprinted genes silenced? S. Tilghman, Cell 96:185 Mechanism- Antisense transcription of unmethylated chromosome blocks sense strand transcription Mechanism- Antisense RNA blocks sense strand transcription
- 37. Prader-Willi Syndrome Normal expression patterns = ON In Prader-Willi deletion, maternal and paternal copies are silent In Angelman, many genes are activated, but UBE3A is silenced
- 38. Prader-Willi Syndrome Proposed mechanism of SNPRN imprinting In Prader Wille, the Switch Initiator Site (SNSIS) is mutated in paternal chromosome, such that it can’t bind BD RNA Imprintor gene RNA (BD RNA) SNSIS SNRPN gene Female allele Imprintor gene RNA (BD RNA) SNSIS SNRPN gene Male allele Dnmt 1 2 3