Session 1 part 3
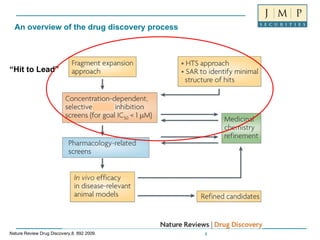
- 1. An overview of the drug discovery process “ Hit to Lead” Nature Review Drug Discovery,8, 892 2009.
- 5. Lead Optimization Nat Rev Drug Disc 2, 369-78, 2003 Medicinal chemist In vivo efficacy is key
- 6. An overview of the drug discovery process
- 7. Medicinal Chemistry Refinement Synthesis of compounds Screen for activity AND/OR Screen against activity AND/OR Screen for ADME Data Analysis (SAR trends) Refinement of criteria Planning Many compounds must be made! What are the strategies used for efficient synthesis? What tools are in the chemists’ synthetic toolbox?
- 9. A case study for library design R. J. Gillespie et al. / Bioorg. Med. Chem. 17 (2009) 6590 – 6605 A diversifiable scaffold with three synthetic handles Facile coupling reactions with commercially available amines create a library to explore space around this position The more reactive chloride can be replaced with various groups through carbon-carbon bond formation The chloride can be substituted with various heteroatoms and groups Straightforward chemistries and commercial reagents allow for rapid diversification Prioritization is necessary
- 10. An overview of the drug discovery process
- 12. Discovery synthesis vs API synthesis: A case study The chosen compound 5 has a m ethyl group added in the last step via a Pd catalyzed reaction as part of a parallel chemistry scheme
- 13. Synthetic scheme for compound 5 as an API W. Hu et al. / Bioorg. Med. Chem. Lett. 17 (2007) 414–418 Methyl group is set early in the synthesis via a cyclization reaction “ Green chemistry”
- 16. Thank you for your attention!
- 18. A structure – toxicity study - A 2A antagonists A2A binding: 2.8 nm A1 binding: 601 nm 3mg/kg p . o . efficacious in vivo for anti-cataleptic activity Molecular Weight: 449.51 log P: 3.33 tPSA: 100.51 hERG inhibition of 81% Maintain potency and selectivity while decreasing hERG % inhibition J. J. Matasi et al. / Bioorg. Med. Chem. Lett. 15 (2005) 3670–3674 J. J. Matasi et al. / Bioorg. Med. Chem. Lett. 15 (2005) 3675–3678
- 19. Natural Products as Drug Starting Points Frank E. Koehn 6 th Drug Discovery for Neurodegeneration February 13 th , 2012 New York, NY
- 23. Screening for Lead Generation Target Compounds Biochemical HTS (Single target) Target-compound binding Phenotypic Screening (many targets) NP chemical Library Phenotypic response New target & mechanism Cell
- 24. Screening and Natural Products Library Design minutes ABSORBANCE 0 2 4 6 8 10 12 14 16 18 20 22 24 26 28 30 32 34 . Media components polar metabolites & biopolymers Lipids, fatty acids non-polar biopolymers Crude Extract Library Fractions/extract Library size per culture Low Assay interferences High Sample prep Low Redundancy High Hit identification Slow Sensitivity 10X Pre-fractionated Library 0 2 4 6 8 10 12 14 16 18 20 22 24 26 28 30 32 34 0 Moderate Moderate Moderate Moderate Moderate 100X 2 4 6 8 10 12 14 16 18 20 22 24 26 28 30 32 34 Pure Compound Library Moderate Low High Very Low Rapid 10 Liter Fermentation 100 Liter Fermentation optimized
- 26. The “Druggable” Genome - Hopkins Highly “Druggable” targets, Ro5 leads Disease relevant “Undruggable” biological targets, Beyond Ro5 leads Very Limited Overlap Hopkins, A.L., Groom, C.R. “The druggable genome” Nat. Rev. Drug Discov., 2002, 1(9) 727-30.
- 27. Natural Products are Successful Therapeutics in the Beyond Ro5 Space Selected Orally Active BRo5 Natural Product Drugs NP Lead, year NCEs Indication/MOA MW ClogP HBD HBA Oral Bioavailability Dose Validamycin, 1970 Acarbose, 1990 Voglibose, 1994 Anti-diabetic/glucosidase inhibitor 498 -6.2 13 14 25 mg Midecamycin, 1971 Miocamycin, 1985 Antibacterial/protein synthesis inhibitors 815 3.5 4 16 100% 600 mg Rapamycin, 1974 Sirolimus, 1999 Everolimus, 2004 Zotarolimus, 2005 Temsirolimus, 2007 Immune suppression/mTOR 914 7.0 3 14 20% 2 mg Cyclosporine A, 1975 Cyclosporine, 1983 Immune suppression /IL-2 inhibitor 1203 14.4 5 23 30% 25 mg Lipstatin, 1975 Orlistat, 1987 Obesity/Lipase inhibitor 492 7.6 1 6 120 mg Avermectin B1a, 1979 Ivermectin, 1987 Antiparasitic/Glutamate-gated chloride channel 873 5.1 3 14 100% 3 mg FK506, 1984 Tacrolimus, 1993 Immune suppression/T-lymphocyte activation inhibitor 804 5.8 3 13 20% 1 mg Myriocin Gilenya, 2010 Multiple sclerosis/S1P1 inhibitor 402 2.8 6 7 93% 0.5 mg
- 28. Recent Synthetic Natural Product Derived Drugs Myriocin Mycelia sterilia Fingolimod Halichondrin B Halichondria okadai Eribulin
- 29. PKS Engineering of Rapamycin 1) Gregory, M.A. and Leadlay, P.F. et al., Angew. Chem. Int. Ed. 2005, 44, 4757-4760. 2) Gregory, M. A. and Leadlay, P.F. et al., Org. & Biomol. Chem. 2006, 4, 3565-3568. rapamycin X X methylation and oxidation Pipecolate Incorporating Enzyme
- 32. Distinct Sequences Often Adopt Very Similar Folds Superposition of 3 proteins of similar structure but distinct sequences. 1 -Isomerase from Rhodopseudomonas palustris 2 - B chain of limonene-1,2-epoxide hydrolase from Rhodococcus erythropolis 3 - Putative polyketide cyclase from Acidithiobacillus ferrooxidans a) 1 and 2 b) 2 and 3 c) 1 and 3 <20% sequence identity in aligned regions Regions of overlap in protein 1 Regions of overlap in protein 2 A- Proteins with virtually identical structure and little or no sequence similarity Current Opinion in Structural Biology 2009, 19:312–320, J Biol Chem 2009, 284:992-999 B- Proteins with high sequence similarity and no structure similarity Arl2 (BART) from Homo sapiens and ADP-ribosylation factor-like protein 2-binding protein from Danio rerio – 72%
- 35. Polyketide Immunophilin Ligand Family Salituro, G. et. al., Tet. Lett., 1995 , 36(7), 997-1000 Summers, M.Y.; Leighton, M.; Liu, D.; Pong, K.; Graziani, E.I., J. Antibiot., 2006 , 59(3), 184-189.
Hinweis der Redaktion
- Untreated cortical neurons (overlay of green (Neurofilament), red (TUJ-1), and blue (Hoechst)). Cortical neurons treated with WAY-265920 .
- mention hot spots, etc.)