Chapter 5 -repair or radiation damage and dose-rate effect - jtl
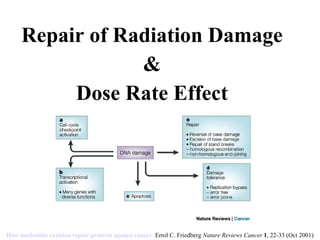
- 1. Repair of Radiation Damage & Dose Rate Effect How nucleotide excision repair protects against cancer Errol C. Friedberg Nature Reviews Cancer 1 , 22-33 (Oct 2001)
- 2. DNA Single Strand Repair
- 4. Base Excision Repair 1) U is 1 st removed by a glycosylase/DNA lyase 2) AP endonuclease then removes the sugar residue 3) Replacement w/correct nucleotide via DNA polymerase f3, 4) DNA ligase III-XRCC1-mediates ligation. * Most common types of DNA damage induced by ionizing radiation are repaired through base excision repair. U represents a putative singlebase mutation
- 10. DNA Double Strand Repair (including HRR and NHEJ) NHEJ is primary means of repairing dsDNA breaks Defects lead to: IR sensitivity, chromosome aberrations
- 11. DNA Double Strand Repair Non Homologous End- Joining (NHEJ) 1) Initiation (End recognition): - Binding of the Ku heterodimer to a dsDNA end 2) End Processing - Recruitment of DNA-dependent protein kinase catalytic subunit, DNAPKcs Artemis activation - Artemis: Endonuclease dependent cleavage of overhangs 3) Synthesis : DNA polymerase 4) Bridging/Joining : XRCC4/DNA ligase IV/XLF complex DNAPK: functions as a regulatory component of NHEJ, potentially facilitating and regulating the processing of the DNA ends. Recruits complex of three proteins, XRCC4, DNA ligase IV and XLF, which carry out the final rejoining step. *Primary means of dsDNA break repair *Error prone* 2/2 in G1 w/no sister chromatid *Important for Ab diversity VDJ recombination
- 15. No (uses template) Yes Sequence Loss Decreases No effect Proliferation Dependent Independent BRCA Dependent Independent ATM G2 G1 Phase of Cell Cycle No Yes Replication Associated HR NHEJ
- 19. AT Cell-cycle checkpoints and cancer Michael B. Kastan and Jiri Bartek Nature 432 , 316-323(18 November 2004) doi:10.1038/nature03097
- 20. ATR
- 21. SCID SCID mice are immunodeficient due to defective DNA-PKcs Such cells are defective in nonhomologous recombination and are extremely radiosensitive. ·SCID humans are also immune deficient and radiosensitive owing to a mutation in Artemis . Cells defective in Artemis are defective in nonohomologous recombination and are radiosensitive .
- 22. SCID & NBS NBS (Nijmegen Breakage Syndrome) is a very rare disorder that results in increased cancer incidence. Cells defective in NBS lack an S phase checkpoint and are radiosensitive. ATLD (AT-Like Disorder) patients are clinically similar to AT patients except that their defect lies in the MRE7 7 gene . Cells from these patients are also sensitive to ionizing radiation.
- 23. Fanconi Anemia FA (Fanconi Anemia) patients are characterized by their hypersensitivity to cross-linking agents . Although fibroblasts derived from these patients are not sensitive to ionizing radiation , tumors arising in these patients are hypersensitive. The reasons for this are currently unknown.
- 25. Potentially Lethal Damage The component of radiation damage that can be modified by manipulation of the postirradiation conditions is known as potentially lethal damage.
- 26. Potentially Lethal Damage Potentially lethal damage repair can occur if cells are prevented from dividing for 6 hours or more after irradiation ; this is manifest as an increase in survival. This repair can be demonstrated in vitro by keeping cells in saline or plateau phase for 6 hours after irradiation and in vivo by delayed removal and assay of animal tumors or cells of normal tissues.
- 29. Sublethal Damage
- 31. Dose Rate Effect
- 33. Dose Rate Effect There is a dose-rate effect, such that at very low dose rates (<0.01 G/min) there is minimal checkpoint activation, probably due to minimal ATM activation. At ~1 Gy/hr, only Late G2 checkpoint is triggered , leading to reassortment of cells at the G2/M interface (and resulting in the inverse dose rate effect). At higher dose rates , cell cycle progression is inhibited at all checkpoints EXAMPLE
- 34. Dose Rate Effect EXAMPLE
- 36. Brachytherapy
- 37. Brachytherapy
- 38. Brachytherapy
- 39. Brachytherapy
- 41. Radiolabeled Ig Therapy
- 64. No (uses template) Yes Sequence Loss Decreases No effect Proliferation Dependent Independent BRCA Dependent Independent ATM G2 G1 Phase of Cell Cycle No Yes Replication Associated HR NHEJ
- 67. DNA Double Strand Repair Non Homologous End- Joining (NHEJ) 1) Initiation (End recognition): - Binding of the Ku heterodimer to a dsDNA end 2) End Processing - Recruitment of DNA-dependent protein kinase catalytic subunit, DNAPKcs Artemis activation - Artemis: Endonuclease dependent cleavage of overhangs 3) Synthesis: DNA polymerase 4) Bridging/Joining: XRCC4/DNA ligase IV/XLF complex DNAPK: functions as a regulatory component of NHEJ, potentially facilitating and regulating the processing of the DNA ends. Recruits complex of three proteins, XRCC4, DNA ligase IV and XLF, which carry out the final rejoining step. *error prone* 2/2 in G1 w/no sister chromatid *Important for Ab diversity VDJ recombination
- 76. DNA Double Strand Repair (including HRR and NHEJ) Depiction of the two main processes for repairing DNA dsb in mammalian cells. In NHEJ, the break is processed and rejoined with the possibility of sequence information being lost and incorrect repair. With HR an intact homolog is used as a template to ensure correct error free repair. : Eur J Pharmacol. Author manuscript; available in PMC 2010 June 25. Eur J Pharmacol. 2009 December 25; 625(1-3): 151–155. Published online 2009 October 14. NHEJ is primary means of repairing dsDNA breaks Defects lead to: IR sensitivity, chromosome aberrations
Hinweis der Redaktion
- Base excision repair BER1, 12, 13, 14, 15, 16, 17 ( a ) is initiated by a class of DNA-repair-specific enzymes — DNA glycosylases — each of which recognizes a single or a small subset of chemically altered or inappropriate bases. For example, an enzyme called uracil DNA-glycosylase specifically recognizes uracil as an inappropriate base in DNA and catalyses hydrolysis of the N -glycosyl bond that links the uracil base to the deoxyribose–phosphate backbone of DNA. Uracil is thus excised from the genome as a free base, leaving a site of base loss in the DNA — an apyrimidinic site in the case of uracil removal, or an apurinic site when a purine is lost. These so-called AP (or abasic) sites are repaired by a further series of biochemical events. How nucleotide excision repair protects against cancer Errol C. Friedberg Nature Reviews Cancer 1 , 22-33 (October 2001)
- a | Nucleotide excision repair (NER) operates on base damage caused by exogenous agents (such as mutagenic and carcinogenic chemicals and photoproducts generated by sunlight exposure) that cause alterations in the chemistry and structure of the DNA duplex . b | Such damage is recognized by a protein called XPC, which is stably bound to another protein called HHRAD23B (R23). c | The binding of the XPC–HHRAD23 heterodimeric subcomplex is followed by the binding of several other proteins (XPA, RPA, TFIIH and XPG). Of these, XPA and RPA are believed to facilitate specific recognition of base damage . TFIIH is a subcomplex of the RNA polymerase II transcription initiation machinery which also operates during NER. It consists of six subunits and contains two DNA helicase activities (XPB and XPD) that unwind the DNA duplex in the immediate vicinity of the base damage. This local denaturation generates a bubble in the DNA, the ends of which comprise junctions between duplex and single-stranded DNA. d | The subsequent binding of the ERCC1–XPF heterodimeric subcomplex generates a completely assembled NER multiprotein complex. e | XPG is a duplex/single-stranded DNA endonuclease that cuts the damaged strand at such junctions 3' to the site of base damage. Conversely, the ERCC1–XPF heterodimeric protein is a duplex/single-stranded DNA endonuclease that cuts the damaged strand at such junctions 5' to the site of base damage. This bimodal incision generates an oligonucleotide fragment 27–30 nucleotides in length which includes the damaged base. f | This fragment is excised from the genome, concomitant with restoring the potential 27–30 nucleotide gap by repair synthesis. Repair synthesis requires DNA polymerases or , as well as the accessory replication proteins PCNA, RPA and RFC . The covalent integrity of the damaged strand is then restored by DNA ligase . g | Collectively, these biochemical events return the damaged DNA to its native chemistry and configuration. ERCC1, excision repair cross-complementing 1; PCNA, proliferating cell nuclear antigen; POL, polymerase; RFC, replication factor C; RPA, replication protein A; TFIIH, transcription factor IIH; XP, xeroderma pigmentosum.
- Mismatch repair MMR 1, 23, 24 ( b ) is a biochemical process dedicated primarily to the excision of nucleotides that are incorrectly paired with the (correct) nucleotide on the opposite DNA strand. Mispairing most frequently (but not exclusively) transpires during DNA replication because of the limited fidelity of the DNA replicative machinery. Hence, the incorrect base occurs in the newly synthesized DNA strand . All cells have specific mechanisms by which they discriminate between newly replicated and parental DNA strands. However, the precise mechanism of strand discrimination in eukaryotic cells is not known . In human cells, the recognition of small loops generated by insertion or deletion of nucleotides, as well as single base mismatches (A:X), is primarily accomplished by a complex called MUTS — a heterodimer of MSH2 and MSH6 . Another heterodimer, MUTS , comprising MSH2 and MSH3, can also operate in the recognition of small loops during MMR. The precise biochemical events subsequent to mismatch recognition in mammalian cells are not well understood, but are believed to involve other heterodimeric complexes, comprising proteins called MLH1, PMS2 and MLH3. As is the case with defective NER, defects in MMR in humans predispose to cancer, in this case primarily to colon cancer 86 but also to uterine 23 , ovarian and gastric cancer.
- Homologous recombination (HR) and non-homologous end-joining (NHEJ) represent two important double strand break (DSB) repair pathways. NHEJ is the major pathway for repairing non-replication associated breaks , which represent most of those induced by ionizing radiation (IR) . By contrast, DSBs arising at the replication fork and some IR-induced DSBs in G2 are repaired by HR . Consequently, cells defective in core NHEJ proteins are exquisitely radiosensitive. Depiction of the two main processes for repairing DNA dsb in mammalian cells. In NHEJ, the break is processed and rejoined with the possibility of sequence information being lost and incorrect repair. With HR an intact homolog is used as a template to ensure correct error free repair. : Eur J Pharmacol. Author manuscript; available in PMC 2010 June 25. Eur J Pharmacol. 2009 December 25; 625(1-3): 151–155. Published online 2009 October 14.
- NHEJ Initiating step in NHEJ is the binding of the Ku heterodimer to a double stranded DNA end . Recruits the DNA-dependent protein kinase catalytic subunit, DNAPKcs , resulting in assembly of the DNAPK complex and activation of its kinase activity. DNAPK: functions as a regulatory component of NHEJ , potentially facilitating and regulating the processing of the DNA ends. Recruits complex of three proteins, XRCC4, DNA ligase IV and XLF, which carry out the final rejoining step. 3) Joining: XRCC4, DNA ligase IV and XLF Ataxia telangiectasia (AT) cells repair most DSBs normally , demonstrating that most NHEJ-dependent DSB repair occurs independently of ataxia telangiectasia mutated (ATM) signalling . On the other hand, cells that lack NHEJ proteins arrest at cell cycle checkpoints. However, a subfraction (15%) of IR-induced DSBs require end-processing by ATM, Artemis and several other DNA damage response (DDR) proteins before repair. Epistasis analysis demonstrates that this process represents a component of NHEJ in non-cycling confluent cells. The precise nature of these DSBs has not been clearly defined, but as they require the nuclease, Artemis, it has been proposed that they represent complex DSBs that need additional processing . Thus, ATM regulates both repair and checkpoint functions . Artemis-defective cells, in contrast to AT cells, display normal cell cycle checkpoint arrest, suggesting that Artemis functions uniquely in the repair arm of ATM signalling.
- restore the functionality of replication forks with DNA doublestrand breaks HR Initiated by strand resection and the recruitment and loading of RAD51 onto single stranded DNA, a process assisted by the BRCA2 protein. Strand invasion creating a D-loop and a Holliday junction then follows. Finally, strand extension by DNA synthesis, typically using the sister chromatid as a template, and resolution of the Holliday junction terminate the HR process. Depending on the cell cycle phase and the nature of the DSB (one-ended versus two-ended DSBs) different subpathways of HR exist (not shown in the figure)
- a | Poly(ADP-ribose) polymerase 1 (PARP1) is shown with its DNA-binding (DBD), automodification (AD) and catalytic domains. The PARP signature sequence (yellow box within the catalytic domain) comprises the sequence most conserved among PARPs. Crucial residues for nicotinamide adenine dinucleotide (NAD+) binding (histidine; H and tyrosine; Y) and for polymerase activity (glutamic acid; E) are indicated. b | Consequences of PARP1 activation by DNA damage. Although not shown to simplify the scheme, PARP1 is active in a homodimeric form116, 117. PARP1 detects DNA damage through its DBD. This activates PARP1 to synthesize poly(ADP) ribose (pADPr; yellow beads) on acceptor proteins, including histones and PARP1. Owing to the dense negative charge of pADPr, PARP1 loses affinity for DNA, allowing the recruitment of repair proteins by pADPr to the damaged DNA (blue and purple circles). Poly(ADP-ribose) glycohydrolase (PARG) and possibly ADP-ribose hydrolase 3 (ARH3) hydrolyse pADPr into ADP-ribose molecules and free pADPr. ADP-ribose is further metabolized by the pyrophosphohydrolase NUDIX enzymes into AMP, raising AMP:ATP ratios, which in turn activate the metabolic sensor AMP-activated protein kinase (AMPK). NAD+ is replenished by the enzymatic conversion of nicotinamide into NAD+ at the expense of phosphoribosylpyrophosphate (PRPP) and ATP. Examples of proteins non-covalently (pADPr-binding proteins) or covalently poly(ADP-ribosyl)ated are shown with the functional consequences of modification (reviewed in Ref. 20). It is important to note that many potential protein acceptors of pADPr remain to be identified owing to the difficulty of purifying pADPr-binding proteins in vivo . PARP inhibitors prevent the synthesis of pADPr and hinder subsequent downstream repair processes, lengthening the lifetime of DNA lesions. ATM, ataxia telangiectasia-mutated; BER, base excision repair; BRCT, BRCA1 carboxy-terminal repeat motif; DNA-PKcs, DNA-protein kinase catalytic subunit; DSB, double-strand break; HR, homologous recombination; NHEJ, non-homologous end joining; NLS, nuclear localisation signal; PPi, inorganic pyrophosphate; SSB, single-strand break; Zn, zinc finger.
- AT = hypersensitive to IR XP = hypersensitive to UV
- AT = hypersensitive to IR
- SCID = Hypersensitive to IR
- Erlotinib or any drug that slows proliferation may result in modification of potentially lethal damage.
- b/c damage is immediate in direct acting IR w/neutrons and indirect w/photons
- Single-track lethal damage is shown to be composed of two components: damage that remains unrepaired in an interval between irradiation and assay, characterized by a very strong dependence on LET, with RBEs up to 20, potentially lethal damage, which is weakly dependent on LET with RBEs < 3. Potentially lethal damage and sublethal damage depend similarly on LET as DNA dsb.
- SUblethal repair occurs w/photons but NOT neutrons
- Neutrons are high LET and thus not affected by fractionation- single track damage causes blobs Multitrack damage causes predominate spurs
- There is a dose-rate effect, such that at very low dose rates (<0.01 G/min) there is minimal checkpoint activation, probably due to minimal ATM activation. At ~1 Gy/hr, only Late G2 checkpoint is triggered, leading to reassortment of cells at the G2/M interface (and resulting in the inverse dose rate effect). At higher dose rates, cell cycle progression is inhibited at all checkpoints There are two distinct G2/M checkpoints ( PMID 11809797 ) A very rapid, dose-independent, ATM-dependent arrest of cells that were in G2 at the time of irradiation (and consequent failure to enter mitosis), which can be assayed using flow cytometry against phosphorylated histone H3 (a marker of mitosis) A slower, dose-dependent, ATM-independent accumulation in G2 of cells which were in earlier stages of the cell cycle at the time of irradiation , which can be assayed using standard propidium iodide flow cytometry
- There are two distinct G2/M checkpoints ( PMID 11809797 ) A very rapid, dose-independent, ATM-dependent arrest of cells that were in G2 at the time of irradiation (and consequent failure to enter mitosis), which can be assayed using flow cytometry against phosphorylated histone H3 (a marker of mitosis) A slower, dose-dependent, ATM-independent accumulation in G2 of cells which were in earlier stages of the cell cycle at the time of irradiation , which can be assayed using standard propidium iodide flow cytometry
- There are two distinct G2/M checkpoints ( PMID 11809797 ) A very rapid, dose-independent, ATM-dependent arrest of cells that were in G2 at the time of irradiation (and consequent failure to enter mitosis), which can be assayed using flow cytometry against phosphorylated histone H3 (a marker of mitosis) A slower, dose-dependent, ATM-independent accumulation in G2 of cells which were in earlier stages of the cell cycle at the time of irradiation , which can be assayed using standard propidium iodide flow cytometry Most cells accumulating in G2/M after IR come from S phase . Schematic representation of cell cycle progression after IR. Characterized checkpoints are labeled with asterisks. ① , G1checkpoint, which is p53 dependent and occurs at restriction point; ② , ATM-dependent S-phase checkpoint, which occurs throughout S phase; ③ , G2/M checkpoint, which is ATM dependent and occurs about 30 min prior to chromosome condensation. The early G2/M checkpoint measures the inhibition of cells that are in G2 at the time of IR from entering mitosis. G2/M accumulation, which is ATM independent , measures the accumulation of cells that were in S phase (or G1) at the time of irradiation. (B) HeLa [ATM(+/+)] and GM9607 [ATM(−/−)] cells were labeled with 30 μM BrdUrd for 30 min and then irradiated with 6 Gy . The top row shows flow-cytometric dot plots of BrdUrd labeling on the ordinate and DNA content on the abscissa before IR and 24 h after IR. The BrdUrd-positive cells (labeled R1) were gated and displayed as DNA content versus cell counts in the bottom row.
- Y-90 : pure beta emitter, average energy 0.94 MeV, tissue penetration 2.5 mm , maximum range 1.1 cm. Half life 64.2 hrs. Clinical use: hepatic microsphere therapy; Zevalin in NHL energetic electron beam is fired at a material like tungsten is due to the deceleration of these electrons as they get deflected by the electrostatic fields of the material’s atoms. This process results in what is called braking radiation - also more commonly known as Bremsstrahlung , a German word meaning the same thing. The electrons in the beam experience the positive and negative electrostatic forces of the material’s atomic nuclei and orbiting electrons. An incoming electron can be deflected from its original direction by these forces and can loose energy which is emitted in the form of an X-ray photon. Typically, the electrons in the beam experience many such interactions before coming to rest. Occasionally, some of them may collide with atomic nuclei, which can give rise to the generation of an X-ray photon whose energy is equal to that of the incoming electron.
- The Zevalin Therapeutic Regimen The ZEVALIN therapeutic regimen is administered on an outpatient basis over the course of 7 to 9 days. The regimen consists of 3 main components:1 Imaging dose: premedication + rituximab + In-111 ZEVALIN Indium scan: gamma camera scan to assess In-111 ZEVALIN biodistribution Therapeutic dose: premedication + rituximab + Y-90 ZEVALIN RIT is a targeted therapeutic approach that combines a monoclonal antibody directed against a specific target antigen with a source of radiation. Although nonradiolabeled monoclonal antibodies can be effective anticancer agents on their own, RIT builds on their cytotoxic potential by adding a radioisotope. RIT permits delivery of a therapeutic dose of radiation directly to the tumor cells. The radiation emitted from the radiolabeled antibody affects not only the antibody-binding cell, but also the neighboring cells. RIT may be beneficial approach for treating non-Hodgkin’s lymphoma (NHL), which is considered to be inherently radiosensitive. The mechanism of action of RIT may be especially beneficial in treating patients with bulky or poorly vascularized tumors.2
- - restore the functionality of replication forks with DNA doublestrand breaks
- - restore the functionality of replication forks with DNA doublestrand breaks
- Homologous recombination (HR) and non-homologous end-joining (NHEJ) represent two important double strand break (DSB) repair pathways. NHEJ is the major pathway for repairing non-replication associated breaks , which represent most of those induced by ionizing radiation (IR) . By contrast, DSBs arising at the replication fork and some IR-induced DSBs in G2 are repaired by HR . Consequently, cells defective in core NHEJ proteins are exquisitely radiosensitive. NHEJ Initiating step in NHEJ is the binding of the Ku heterodimer to a double stranded DNA end. Recruits the DNA-dependent protein kinase catalytic subunit, DNAPKcs, resulting in assembly of the DNAPK complex and activation of its kinase activity. DNAPK: functions as a regulatory component of NHEJ, potentially facilitating and regulating the processing of the DNA ends. Recruits complex of three proteins, XRCC4, DNA ligase IV and XLF, which carry out the final rejoining step. 3) Joining: XRCC4, DNA ligase IV and XLF Ataxia telangiectasia (AT) cells repair most DSBs normally, demonstrating that most NHEJ-dependent DSB repair occurs independently of ataxia telangiectasia mutated (ATM) signalling . On the other hand, cells that lack NHEJ proteins arrest at cell cycle checkpoints. However, a subfraction (15%) of IR-induced DSBs require end-processing by ATM, Artemis and several other DNA damage response (DDR) proteins before repair. Epistasis analysis demonstrates that this process represents a component of NHEJ in non-cycling confluent cells. The precise nature of these DSBs has not been clearly defined, but as they require the nuclease, Artemis, it has been proposed that they represent complex DSBs that need additional processing. Thus, ATM regulates both repair and checkpoint functions. Artemis-defective cells, in contrast to AT cells, display normal cell cycle checkpoint arrest, suggesting that Artemis functions uniquely in the repair arm of ATM signalling. HR Initiated by strand resection and the recruitment and loading of RAD51 onto single stranded DNA, a process assisted by the BRCA2 protein 91 . Strand invasion creating a D-loop and a Holliday junction then follows. Finally, strand extension by DNA synthesis, typically using the sister chromatid as a template, and resolution of the Holliday junction terminate the HR process. Depending on the cell cycle phase and the nature of the DSB (one-ended versus two-ended DSBs) different subpathways of HR exist (not shown in the figure).
- Signal transduction pathways are activated by either ataxia telangiectasia mutated (ATM) or ataxia telangiectasia and RAD3-related (ATR). ATM responds to double strand breaks (DSBs), and evidence suggests an activating role for the NBS1–MRE11–RAD50 (MRN) complex 70 . ATR responds to single stranded regions of DNA, and requires ATR-interacting protein (ATRIP), RPA 71 , the RAD17–RFC2-5 complex, a complex of RAD9–HUS1–RAD1 (the 9–1–1 complex) 72 , TOPBP1 and claspin. The signalling pathways involve the mediator proteins (MDC1, 53BP1, the MRN complex and BRCA1), which amplify the signal, transducer kinases (CHK1 and CHK2) and effector proteins 17 . ATM and ATR show considerable overlap in their phosphorylation substrates but specificity also exists, with CHK1 and CHK2 being specific substrates of ATR and ATM, respectively. Two ATM-dependent G1/S checkpoints have been described. ATM activation by DSBs in G1 leads to CHK2 phosphorylation and subsequent phosphorylation of the phosphatase CDC25A. This increases the ubiquitylation and proteolytic degradation of CDC25A and prevents activating dephosphorylation of CDK2 at Thr14 and Tyr15 59, 73, 74 . As this pathway operates through post-translational modifications, initiation of the G1/S checkpoint occurs soon after damage induction. A second mechanism involves the tumour suppressor p53, which is activated and stabilized by ATM, either directly or indirectly through CHK2, and serves as a transcription factor for the cyclin-dependent kinase (CDK) inhibitor p21 (Refs 49 , 58 , 59 ). This pathway depends on the transcription of p21 and is therefore delayed after ionizing radiation (IR), suggesting that its role lies in the maintenance of the G1/S checkpoint 49, 57, 59, 75 . Collapsed or stalled replication forks in S phase activate ATR, leading to the phosphorylation of CHK1 and the subsequent phosphorylation and proteolysis of CDC25A 59 . This prevents initiation of new replication origins and slows down replication. A second branch of this intra-S phase checkpoint involves the MRN complex, BRCA1, FANCD2 and SMC1 76 (not shown in figure). DSBs in G2 can directly activate ATM, and indirectly, via ATM-dependent strand resection, can lead to ATR activation 64 . Similar to the rapid activation of the G1/S and intra-S phase checkpoints, the G2/M checkpoint is initiated by the phosphorylation of checkpoint kinases (CHK1 and CHK2) and phosphatases (probably CDC25C). This prevents dephosporylation of CDK1–cyclin B, which is required for progression into mitosis 23, 59 . Although p53 has an essential role in p21-dependent G1/S arrest , it promotes but is non-essential for the G2/M checkpoint . Roles include an affect on the maintenance of checkpoint arrest and on CDK1–cyclin B activity through transcriptional activation and regulation of GADD45 and 14-3-3
- - restore the functionality of replication forks with DNA doublestrand breaks
