2014 agbt giab data integration poster 140206
•Als PPTX, PDF herunterladen•
0 gefällt mir•1,278 views
Melden
Teilen
Melden
Teilen
Empfohlen
Empfohlen
Use of CEDAR Technology for Ontology-based Submission of Biomedical Data to the NCBI
Use of CEDAR Technology for Ontology-based Submission of Biomedical Data to ...

Use of CEDAR Technology for Ontology-based Submission of Biomedical Data to ...Syed Ahmad Chan Bukhari, PhD
Phylogenetics: Making publication-quality tree figuresPhylogenetics: Making publication-quality tree figures

Phylogenetics: Making publication-quality tree figuresBioinformatics and Computational Biosciences Branch
Weitere ähnliche Inhalte
Was ist angesagt?
Use of CEDAR Technology for Ontology-based Submission of Biomedical Data to the NCBI
Use of CEDAR Technology for Ontology-based Submission of Biomedical Data to ...

Use of CEDAR Technology for Ontology-based Submission of Biomedical Data to ...Syed Ahmad Chan Bukhari, PhD
Phylogenetics: Making publication-quality tree figuresPhylogenetics: Making publication-quality tree figures

Phylogenetics: Making publication-quality tree figuresBioinformatics and Computational Biosciences Branch
Was ist angesagt? (20)
171114 best practices for benchmarking variant calls justin

171114 best practices for benchmarking variant calls justin
tools for reproducible research in an increasingly digital world

tools for reproducible research in an increasingly digital world
IRIDA: Canada’s federated platform for genomic epidemiology 

IRIDA: Canada’s federated platform for genomic epidemiology
Use of CEDAR Technology for Ontology-based Submission of Biomedical Data to ...

Use of CEDAR Technology for Ontology-based Submission of Biomedical Data to ...
Tools and approaches for data deposition into nanomaterial databases

Tools and approaches for data deposition into nanomaterial databases
How Can We Make Genomic Epidemiology a Widespread Reality? - William Hsiao

How Can We Make Genomic Epidemiology a Widespread Reality? - William Hsiao
How giab fits in the rest of the world mdic somatic reference samples

How giab fits in the rest of the world mdic somatic reference samples
2016 ACS Semantic Approaches for Biochemical Knowledge Discovery

2016 ACS Semantic Approaches for Biochemical Knowledge Discovery
Phylogenetics: Making publication-quality tree figures

Phylogenetics: Making publication-quality tree figures
Ähnlich wie 2014 agbt giab data integration poster 140206
Ähnlich wie 2014 agbt giab data integration poster 140206 (20)
GIAB-GRC workshop oct2015 giab introduction 151005

GIAB-GRC workshop oct2015 giab introduction 151005
GIAB Benchmarks for SVs and Repeats for stanford genetics sv 200511

GIAB Benchmarks for SVs and Repeats for stanford genetics sv 200511
Using accurate long reads to improve Genome in a Bottle Benchmarks 220923

Using accurate long reads to improve Genome in a Bottle Benchmarks 220923
Genome in a bottle for ashg grc giab workshop 181016

Genome in a bottle for ashg grc giab workshop 181016
Genome in a Bottle- reference materials to benchmark challenging variants and...

Genome in a Bottle- reference materials to benchmark challenging variants and...
Mehr von GenomeInABottle
Mehr von GenomeInABottle (20)
GIAB Technical Germline Benchmark roadmap discussion

GIAB Technical Germline Benchmark roadmap discussion
Ga4gh 2019 - Assuring data quality with benchmarking tools from GIAB and GA4GH

Ga4gh 2019 - Assuring data quality with benchmarking tools from GIAB and GA4GH
GIAB GRC Workshop ASHG 2019 Billy Rowell Evaluation of v4 with CCS GATK

GIAB GRC Workshop ASHG 2019 Billy Rowell Evaluation of v4 with CCS GATK
GRC GIAB Workshop ASHG 2019 Small Variant Benchmark

GRC GIAB Workshop ASHG 2019 Small Variant Benchmark
Genome in a Bottle - Towards new benchmarks for the “dark matter” of the huma...

Genome in a Bottle - Towards new benchmarks for the “dark matter” of the huma...
How giab fits in the rest of the world seqc2 tumor normal

How giab fits in the rest of the world seqc2 tumor normal
New data from giab genomes intro and ultralong nanopore

New data from giab genomes intro and ultralong nanopore
2014 agbt giab data integration poster 140206
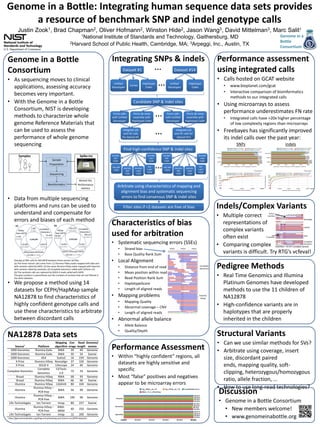
- 1. Genome in a Bottle: Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls Justin 1, Zook Brad 2, Chapman Oliver 2, Hofmann Winston 2, Hide Jason 3, Wang David 3, Mittelman 1National Institute of Standards and Technology, Gaithersburg, MD 2Harvard School of Public Health, Cambridge, MA; 3Arpeggi, Inc., Austin, TX 1 Integrating SNPs & indels Genome in a Bottle Consortium • As sequencing moves to clinical applications, assessing accuracy becomes very important. • With the Genome in a Bottle Consortium, NIST is developing methods to characterize whole genome Reference Materials that can be used to assess the performance of whole genome sequencing Samples Spike-ins Sample Preparation Unified Genotyper Force calls with Unified Genotyper • Data from multiple sequencing platforms and runs can be used to understand and compensate for errors and biases of each method Force de novo assembly with Haplotype Caller … Unified Genotyper Haplotype Caller Force calls with Unified Genotyper … Force de novo assembly with Haplotype Caller NA12878 Data sets • • www.bioplanet.com/gcat Interactive comparison of bioinformatics methods to our integrated calls • Using microarrays to assess performance underestimates FN rate • Integrated calls have >20x higher percentage of low complexity regions than microarrays SNPs indels Find high-confidence SNP & indel sites HomRef SNP VQSR HomRef indel VQSR HomVar SNP VQSR HomVar indel VQSR Het indel VQSR … HomRef SNP VQSR Het SNP VQSR HomRef indel VQSR HomVar SNP VQSR HomVar indel VQSR Het indel VQSR Arbitrate using characteristics of mapping and alignment bias and systematic sequencing errors to find consensus SNP & indel sites Indels/Complex Variants Filter sites if <2 datasets are free of bias • Multiple correct representations of complex variants often exist • Comparing complex CAGTGA > TCTCT complex variant variants is difficult. Try RTG’s vcfeval! Characteristics of bias used for arbitration • • • We propose a method using 14 datasets for CEPH/HapMap sample NA12878 to find characteristics of highly confident genotype calls and use these characteristics to arbitrate between discordant calls Performance assessment using integrated calls • Freebayes has significantly improved its indel calls over the past year: Integrate UG and HC calls for dataset #11 • Systematic sequencing errors (SSEs) Overlap of SNP calls for NA12878 between three variant call files. (a) The three variant calls come from: (1) Illumina HiSeq reads mapped with bwa and with variants called by GATK; (2) the same Illumina HiSeq reads mapped with bwa but with variants called by samtools; (3) Complete Genomics called with CGTools 2.0. (b) The samtools calls are replaced by SOLiD 4 reads called with GATK. The gray numbers in parentheses are the numbers of variants that are not filtered in the other datasets. Genome in a Bottle Consortium • Calls hosted on GCAT website Haplotype Caller Integrate UG and HC calls for dataset #1 Sequencing Variant list, Performance metrics Cortex Dataset #14 Candidate SNP & indel sites Het SNP VQSR Bioinformatics … … Dataset #1 Marc Salit1 Strand bias Base Quality Rank Sum • Local Alignment • • • • • • Mapping problems • • • Complete Genomics Distance from end of read Mean position within read Read Position Rank Sum HaplotypeScore Length of aligned reads Illumina HiSeq Mapping Quality Abnormal coverage – CNV Length of aligned reads • Abnormal allele balance • • Allele Balance Quality/Depth Performance Assessment • Within “highly confident” regions, all datasets are highly sensitive and specific • Most “false” positives and negatives appear to be microarray errors Pedigree Methods • Real Time Genomics and Illumina Platinum Genomes have developed methods to use the 11 children of NA12878 • High-confidence variants are in haplotypes that are properly inherited in the children Structural Variants • Can we use similar methods for SVs? • Arbitrate using coverage, insert size, discordant paired ends, mapping quality, softclipping, heterozygous/homozygous ratio, allele fraction, … • How to use long-read technologies? Discussion a http://genomeinabottle.org/blog-entry/existing-and-future-na12878-datasets. • Genome in a Bottle Consortium • New members welcome! • www.genomeinabottle.org
