Restriction Mapping
- 1. Restriction Mapping SUNIL BHANDARI BALKUMARI COLLEGE
- 2. Restriction enzymes: • Restriction enzymes are one of the most important tools in the recombinant DNA technology. • These are DNA cutting enzyme present in bacteria (prokaryotes) that recognizes the specific sites in the DNA, called restriction sites and make a DNA double strand break at or near recognition sites. • After cutting, the newly produced DNA ends will have either a blunt ended or sticky (staggered) end.
- 3. Restriction Enzyme/Endonuclease “Also known as molecular scissor” • Type I enzymes: They are complex multi subunit combination restriction and modification enzymes that cut DNA at random far from their recognition sequences • Type II enzymes: Cut DNA at defined positions close to or within their recognition sequences • Type III enzymes: They cleave outside of their recognition sequences and require two such sequences in opposite orientations within the same DNA molecule to accomplish cleavage they rarely give complete digests • Type IV enzymes: Recognize modified, typically methylated DNA and cleave at that region
- 4. Example of restriction enzymes and their corresponding restriction sits
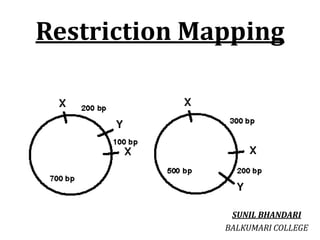
- 5. Restriction Mapping • Restriction mapping is a method used to map an unknown segment of DNA by breaking it into pieces and then identifying the locations of the breakpoints. This method relies upon the use of proteins called restriction enzymes, which can cut, or digest, DNA molecules at short, specific sequences called restriction sites After a DNA segment has been digested using a restriction enzyme, the resulting fragments can be examined using a laboratory method called gel electrophoresis, which is used to separate pieces of DNA according to their size. • One common method for constructing a restriction map involves digesting the unknown DNA sample in three ways. Here, two portions of the DNA sample are individually digested with different restriction enzymes, and a third portion of the DNA sample is double digested with both restriction enzymes at the same time • Next, each digestion sample is separated using gel electrophoresis, and the sizes of the DNA fragments are recorded. The total length of the fragments in each digestion will be equal However, because the length of each individual DNA fragment depends upon the positions of its restriction sites, each restriction site can be mapped according to the lengths of the fragments.
- 6. • The information from the double digestion is particularly useful for correctly mapping the sites. The final drawing of the DNA segment that shows the positions of the restriction sites is called a restriction map • It is possible to determine the position of restriction sites on a DNA fragment by digesting it with various restriction enzymes, singly and in combination, and analyzing the fragment sizes obtained. • Restriction mapping used to be a key procedure for characterizing a cloned fragment of DNA, but this is now more easily done by DNA sequencing. • Nevertheless, analysis of restriction sites (or fragment sizes) is still useful, for example in comparing the chromosomal organization of different strains.
- 9. Assignment:01 Consider, a 4kb DNA is digested with two restriction enzymes (HindIII and BamHI).The digested product is run through gel electrophoresis and DNA bands of following size is obtained. Draw the restriction map representation following digestion. Ans: See last page
- 10. Application of Restriction Mapping: 1. Identification of restriction sites 2. Insertion and deletion study of a gene 3. Insert analysis during cloning 4. Verification of the size of the insert during cloning 5. Mutation studies
- 11. Thank you…
