Proposal for Protein-DNA Mapping using AFM for Lab on a Chip
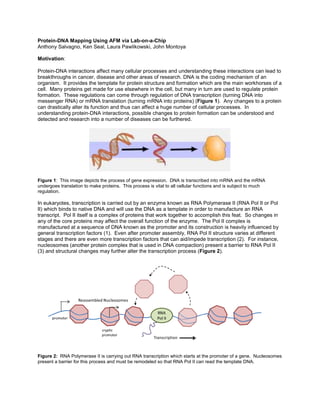
- 1. Protein-DNA Mapping Using AFM via Lab-on-a-Chip Anthony Salvagno, Ken Seal, Laura Pawlikowski, John Montoya Motivation: Protein-DNA interactions affect many cellular processes and understanding these interactions can lead to breakthroughs in cancer, disease and other areas of research. DNA is the coding mechanism of an organism. It provides the template for protein structure and formation which are the main workhorses of a cell. Many proteins get made for use elsewhere in the cell, but many in turn are used to regulate protein formation. These regulations can come through regulation of DNA transcription (turning DNA into messenger RNA) or mRNA translation (turning mRNA into proteins) (Figure 1). Any changes to a protein can drastically alter its function and thus can affect a huge number of cellular processes. In understanding protein-DNA interactions, possible changes to protein formation can be understood and detected and research into a number of diseases can be furthered. Figure 1: This image depicts the process of gene expression. DNA is transcribed into mRNA and the mRNA undergoes translation to make proteins. This process is vital to all cellular functions and is subject to much regulation. In eukaryotes, transcription is carried out by an enzyme known as RNA Polymerase II (RNA Pol II or Pol II) which binds to native DNA and will use the DNA as a template in order to manufacture an RNA transcript. Pol II itself is a complex of proteins that work together to accomplish this feat. So changes in any of the core proteins may affect the overall function of the enzyme. The Pol II complex is manufactured at a sequence of DNA known as the promoter and its construction is heavily influenced by general transcription factors (1). Even after promoter assembly, RNA Pol II structure varies at different stages and there are even more transcription factors that can aid/impede transcription (2). For instance, nucleosomes (another protein complex that is used in DNA compaction) present a barrier to RNA Pol II (3) and structural changes may further alter the transcription process (Figure 2). Figure 2: RNA Polymerase II is carrying out RNA transcription which starts at the promoter of a gene. Nucleosomes present a barrier for this process and must be remodeled so that RNA Pol II can read the template DNA.
- 2. Because all cell life and functions take root in transcription, changes to the process can lead to some serious problems. Simple mutations to DNA sequence can alter protein binding sites which can affect a large number of events (see above). Similar sequence alterations will affect protein structure, or could even alter transcription length. Protein modifications of DNA binding proteins have been linked to various forms of cancer (4), neurological disease (5), and heart disease (6), just to name a few. The intended research is to map protein locations on DNA of healthy functional cells and compare them to protein locations from unhealthy cells (of various causes). Structural changes may lead to different binding sites which will affect protein making processes (transcription). We hypothesize that we can map protein-DNA interaction locations (binding sites) and could detect structural changes of proteins to add further insight to interactions between protein and DNA. Approach: Detection of proteins on a DNA sequence will be achieved through the use of an AFM tip coated with antibodies. In order for our bodies to fight infection properly there is a sort of surveillance system that properly tags foreign objects. When a nonnative substance (proteins, enzymes, molecules, etc.) enters a vertebrate body the foreign substance is tagged and neutralized with the use of immunoglobulins, or antibodies. For each foreign particle (antigens) there is a specific antibody that can target that object. Antibodies all have the same basic subunits. They are made up of two heavy chains and two light chains and this basic design (Figure 3) allows for more complex structures to form (monomers, dimers, etc.). The most important feature of antibody design is known as the hypervariable region. This region can be changed to complement any antigen and is the location where antigens can be bound. The interaction between an antigen and its antibody is highly specific and allows an antibody to precisely label only one antigen. It is this interaction mechanism that we will utilize in this study. Figure 3: This is the simple structure of an antibody, which is made up of two heavy chains and two short chains. Antigens can bind to the antibody via the hypervariable region. This interaction can be envisioned similar to a lock and key setup. Antibodies are readily available for purchase but in some cases they may have to be constructed. In today's bioengineered world antibody acquisition is significantly easier than in years past. Antigen
- 3. insertion into mammals will yield antibody construction. Simple extraction and isolation from the blood of an animal will provide an experimenter with polyclonal antibodies. Polyclonal antibodies will all bind to the same antigen, but are of differing structure. In order to obtain a more uniform antibody family, monoclonal antibodies will need to be produced by cloning a single immune cell that produces the proper antibody. AFM has previously been used to characterize DNA for different applications. Previous groups have demonstrated its versatility in mapping exonuclease activicies of DNA (7). In this work they did not use a functionalized AFM tip but a regular tip in tapping mode. This allowed the experimenters to map out the contours of the exonuclease. Similar to this work was the work involving the interaction between thalidomide and DNA (8). Like in the previous application, this work just studied the topography of the substance. In addition to unfunctionalized AFM tips, others have modified the tip for different experiments. The work of Meister et al. demonstrated the use of AFM tips as a way to inject substances into nanochannels (9). The AFM tip was modified with a drill to allow for a small channel of liquid to flow through the tip into the nanochannels (Figure 4). This device successfully demonstrated the use of AFM in nanofluidic application. Figure 4: Schematic of a modified AFM tip. The tip has been hallowed out and drilled at the end to allow liquid to flow through the tip onto a device. One form of graphing different materials on tips is the use of an electron beam (10). This was used to attach carbon nanotubes to the tips. The tubes were placed near the tips, and then guided onto the tips. The electron beam was then used to modify the size of the tips. This method was able to sharpen the edge of the AFM tips allowing them to be more accurate in their measurements. For this work, an AFM tip is fabricated using conventional MEMS device fabrication techniques with a built-in capacitor. Once the AFM chip is fabricated, the tip is modified with antibodies. When proteins are detected, the AFM tip deflects which causes a change in voltage on the built-in capacitor. This is due to the interactions between the proteins and the antibodies. When the protein comes in contact with its
- 4. respective antibody it will cause a deflection in the AFM tip again allowing the change in voltage to be measure. As described above the AFM tip must be modified with antibodies so that the voltage change can be measure. To do this, the antibodies must be attached to the tip. The tips should also be capable of attaching to many different types of antibodies so that more proteins can be analyzed. There have been several different groups that have been able to modify their tips and attach different products to them. In one application the tips were exposed to UV light to prevent contamination of any organic element. After the tips have been sanitized they are places in a plasmid solution with the desired encoding material. They are then allowed to incubate for a long period of time to allow for the material to adhere to the surfaces(11). This method would be the most beneficial to the experiment in question. It will allow for the antibodies to attach to the surface of the AFM tip. Unlike the other methods mentioned above, this design allows for a low cost modification of the AFM tips. Since the main focus of this device will include a capacitance measurement, the modified tips will not be used in a typical AFM application. Instead they will be incorporated directly into the devices themselves. Process 1: AFM Device Fabrication, Wafer 1 Process 1 represents the fabrication of an atomic force microscope (AFM) tip made from SU-8 photoresist. SU-8 photoresist is a common photoresist used to fabricate permanent MEMS structures because it is a structurally reliable material. This AFM tip will also include a built-in capacitor made from gold to measure minute changes in probe height. The first few steps are designed to fabricate the shape of the AFM tip in a silicon wafer using well known photolithography techniques. An anisotropic etch is first performed to define the cantilever portion of the AFM probe. Steps 1 and 2 demonstrate this process (Figure 5a, b). After the anisotropic etch occurs, a selective silicon plane etch is then performed to form a sharp AFM tip to probe the small features of DNA. This is shown in step 3 of the (Figure 5c). SU-8 works in a similar fashion as standard photoresist, except that its chemical composition is designed to form structurally stable “glass” after a high temperature hard bake process. Step 4 demonstrates the deposition of SU-8 (Figure 5d). A final modification step includes the deposition of gold onto the top part of the device. This will define the bottom part of the capacitor. This step is illustrated as step 5 (Figure 5e).
- 5. Figure 5: Initial AFM fabrication steps to define an SU-8 tip and cantilever with an electrode acting as the bottom portion of a capacitor. Process 1: AFM Device Fabrication, Wafer 2 A second wafer is processed to define the top portion of the AFM capacitor as well as a protective cover to the AFM probe. To begin this process, a deep well is anisotropically etched into a silicon wafer (Figure 6a). Once the second wafer is etched, a layer of silicon dioxide is deposited to protect any the electrical contacts from electrical noise that might result from standard (unintentionally doped) silicon wafers (Figure 6b). Gold metal is deposited to define two electrodes. One electrode will act as a bottom capacitor contact, and the other electrode will act as the top part of the capacitor (Figure 6c). Indium metal is used to help bond “wafer 1” with “wafer 2” because it melts at a low temperature (156 degrees Celsius). A low temperature bond is desirable because high temperatures could cause damage to the chip (Figure 6d).
- 6. Figure 6: Fabrication steps that define the AFM cover and capacitor contact electrodes. Process 1: AFM Device Fabrication, Wafer 1 & 2 Wafer 1 and 2 are bonded together by applying pressure at the melting temperature of indium (Figure 7a). The AFM device fabrication process is completed by releasing the AFM probe from silicon with an isotropic wet etch (Figure 7b). Figure 7: Final processing steps to define the AFM tip and capacitor electrodes. Process 2: DNA Nanofluidic Channel Fabrication Process 2 represents the fabrication process of the nanofluidic channels. This is done with standard nano imprint lithography (21). The process is started by first coating a silicon wafer with PMMA polymer (Figure 8a). A nanofluidic channel template is used to imprint the PMMA polymer with a defined structure. The template is made with standard anisotropic etch techniques, with the nano features further defined with a focused ion beam (FIB) machine (Figure 8b). Next, the nanofluidic channels are sealed with a top layer of PMMA polymer. This is done by coating a layer of lift of resist (LOR) onto a dummy wafer with a layer of PMMA polymer on top. The wafer with the imprinted nanofluidic channels is bonded with the dummy wafer by applying heat and pressure. The dummy wafer is removed by exposing the structure to commercial developer that reacts only with LOR material (Figure 8c). In order to allow the AFM probe access to DNA located into the nanofluidic channel, a hole must be defined into the PMMA cover layer. This hole is defined by performing a shallow anisotropic etch (Figure 8d). Gold electrical contacts are deposited to make contact to the top and bottom
- 7. part of the capacitor (Figure 8e). Indium is used again to bond the AFM chip with the nanofluidic chip (Figure 8f). Figure 8: Processing steps used to define the nanofluidic channels and electrodes used to make contact with the electrodes on the AFM chip. End Process: Flip-Chip AFM Chip with Nanofluidic Chip The AFM chip and the nanofluidic chip are brought together and bonded with a flip-chip bonding machine. Figure 9: Side view completed protein bound DNA detector made from standard photolithography fabrication techniques.
- 8. Figure 10: Top view of completed protein bound DNA detector made from standard photolithography fabrication techniques. Figure 11: Front side view of completed protein bound DNA detector made from standard photolithography fabrication techniques. Proposed Device Operation: Proteins within the channel will interact with their corresponding antibodies. This will cause the AFM tip to be deflected. When the AFM tip deflects, the distance between the top capacitor electrode and the bottom capacitor electrode changes causing a change in voltage. A change in voltage occurs because of the relation V=(Distance)*(Electric Field). This change in voltage determines where a protein is located within the DNA strand. By creating multiple chips with different antibodies and connecting them in sequence, the location of many proteins can be determined.
- 9. Expected Results: The use of antibodies for protein detection is nothing new and their functions have paved the way for various applications in the field of biosensors (12). In initial tests we expect to purchase required antibodies to test the chip. The chip will be designed so that labs wishing to study a particular protein can use their own purified antibodies and easily affix them to the "tip" for experimentation. For more complex studies we would require collaboration with a lab that can provide us with the appropriate antibodies. Although the design of our chip is thorough, potential problems could still arise. Changes in the flow of buffer solution as well as system pressure will lead to "natural" fluctuations of the AFM tip. This background noise should be normalized to ensure proper decrease in voltage upon antibody antigen interaction. In order for the experiment to be successful, it is necessary the chip mimic the human body's internal cellular environment throughout. Maintaining correct ion concentrations and pH is crucial. Inaccurate results could be detected should any deviations occur. Once the protein bound DNA fragments of interest are inserted into the chip, capillary action will be the main driving force to guide the DNA protein complex through the nanofluidic channels. If capillary action is problematic, an electroosmotic microfluidic pump could easy be incorporated to the chip design (13). Once the protein bound DNA is successfully inserted, the researcher has no way of determining the orientation of the DNA fragment (i.e. is the head 3' or 5'). It is crucial the researcher know the approximate length in base pairs, as well as the approximate location of the protein recognition sequence on the DNA. If the above are known, the DNA protein complex is ready for detection. As the DNA protein complex travels through the chip on its way to the detector, it encounters its first bottleneck (Figure 8d). Successful insertion into the detection channel is not well understood and potentially problematic. This design relies on capillary action as its main driving force. While the DNA protein complex is traveling through the chip, it is essential the complex enter the detection channel in a linear head-to-tail fashion since the detection channel is only wide enough to accommodate one DNA protein complex at a time. Should a DNA protein complex be forced through the middle of a strand, this strand would act as a molecular plug blocking the detection channel. After a successful linear head-to-tail insertion of the DNA protein complex, the antibody labeled AFM tip is poised for detection. Theoretically, the DNA protein complex should pass through the nano-fluidic channel via capillary action, and once the antibody recognizes its antigen (in this case, the DNA bound protein), the capacitor will detect a drop in voltage (Figure 9) thus detecting the protein of interest. In order for the antibody coated AFM tip to successfully detect its antigen via a drop in voltage, the following criteria must be met. First, as mentioned earlier, the DNA protein complex must enter the detection channel in a head-to-tail fashion. Should the DNA protein complex fold back upon itself and travel through the detection channel at greater than twice the height of a normal DNA protein complex, the detector would register a sharp increase in voltage. Provided the folded DNA protein complex does not block the detection channel, the results would be ignored. Secondly, after successful entry to the detection channel, it is possible for the antibody to bind to its antigen thus blocking the detection channel. Although the proposed design has potential flaws, the possibility for rapid protein bound DNA detection is useful (5,6). This DNA detector uses simple photolithography techniques allowing for low cost quick fabrication of the devices. The proposed lab-on-a-chip design could save researchers precious time and money. This design opens up opportunities for new developments in DNA detection for a broader range of genetic diseases.
- 10. References 1. Juven-Gershon, T., Hsu, J. Y., Theisen, J. W. & Kadonaga, J. T. The RNA polymerase II core promoter — the gateway to transcription. Curr. Opin. Cell Biol., 2008, 20, 253–259 . 2. Fuda, N. J.; Ardehali, M. B.; Lis, J. T., Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 2009, 461 (7261), 186-192. 3. Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal Fluctuations Govern the Transcription Dynamics of RNA Polymerase II. Science, 2009, 325 (626). 4. Yoo Y, Hayashi M, Christensen J, Huang LE. An essential role of the HIF-1alpha-c-Myc axis in malignant progression. Annals of the New York Academy of Sciences 2009, 1177, 198-204. 5. Borrelli E, Chambon P. Control of transcription and neurological diseases. Molecular psychiatry 1999, 4 (2), 112-4. 6. Schott, JJ, et al. Congenital Heart Disease Caused by Mutations in the Transcription Factor NKX2-5. Science 1998, 281 (5373), 108. 7. Hori, K.; Takahashi, T.; Okada, T., The measurement of exonuclease activities by atomic force microscopy. European Biophysics Journal with Biophysics Letters 1998, 27 (1), 63-68. 8. Oliveira, S. C. B.; Chiorcea-Paquim, A. M.; Ribeiro, S. M.; Melo, A. T. P.; Vivan, M.; Oliveira-Brett, A. M., In situ electrochemical and AFM study of thalidomide-DNA Interaction. Bioelectrochemistry 2009, 76 (1-2), 201-207. 9. Meister, A.; Gabi, M.; Behr, P.; Studer, P.; Voros, J.; Niedermann, P.; Bitterli, J.; Polesel-Maris, J.; Liley, M.; Heinzelmann, H.; Zambelli, T., FluidFM: Combining Atomic Force Microscopy and Nanofluidics in a Universal Liquid Delivery System for Single Cell Applications and Beyond. Nano Letters 2009, 9 (6), 2501-2507. 10. Martinez, J.; Yuzvinsky, T. D.; Fennimore, A. M.; Zettl, A.; Garcia, R.; Bustamante, C., Length control and sharpening of atomic force microscope carbon nanotube tips assisted by an electron beam. Nanotechnology 2005, 16 (11), 2493-2496. 11. Cuerrier, C. M.; Lebel, R.; Grandbois, M., Single cell transfection using plasmid decorated AFM probes. Biochemical and Biophysical Research Communications 2007, 355 (3), 632-636. 12. Conroy, P. J.; Hearty, S.; Leonard, P.; O'Kennedy, R. J., Antibody production, design and use for biosensor-based applications. Seminars in Cell & Developmental Biology 2009, 20 (1), 10-26. 13. Glawdel, T.; Ren, C. L., Electro-osmotic flow control for living cell analysis in microfluidic PDMS chips. Mechanics Research Communications2009, 36 (1), 75-81. 14. Cui, Y.; Wei, Q. Q.; Park, H. K.; Lieber, C. M., Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science 2001, 293 (5533), 1289-1292. 15. Hahm, J.; Lieber, C. M., Direct ultrasensitive electrical detection of DNA and DNA sequence variations using nanowire nanosensors. Nano Letters2004, 4 (1), 51-54. 16. MacBeath, G.; Schreiber, S. L., Printing proteins as microarrays for high-throughput function determination. Science 2000, 289 (5485), 1760-1763. 17. Schadt, E. E., Molecular networks as sensors and drivers of common human diseases. Nature 2009, 461 (7261), 218-223. 18. Star, A.; Gabriel, J. C. P.; Bradley, K.; Gruner, G., Electronic detection of specific protein binding using nanotube FET devices. Nano Letters2003, 3 (4), 459-463. 19. Whang, D.; Jin, S.; Wu, Y.; Lieber, C. M., Large-scale hierarchical organization of nanowire arrays for integrated nanosystems. Nano Letters2003, 3 (9), 1255-1259. 20. Xia, D. Y.; Gamble, T. C.; Mendoza, E. A.; Koch, S. J.; He, X.; Lopez, G. P.; Brueck, S. R. J., DNA transport in hierarchically-structured colloidal-nanoparticle porous-wall nanochannels. Nano Letters 2008, 8 (6), 1610-1618. 21. L. Jay Guo; Xing Cheng; Chia-Fu Chou, Fabrication of Size-Controllable Nanofluidic Channels by Nanoimprinting and Its Application for DNA Stretching. Nano Letters 2004, 4 (1), 69-73.
